Technological Developments
One of the main challenges for research lies in the computer-assisted integrative study of large and increasingly complex combinations of data. The preservation of the materials and methods of such computational experiments with clear annotations is essential for understanding an experiment, and this is increasingly recognized in the bioinformatics and astronomy community. Our assumption is that offering means of digital, structured aggregation and annotation of the objects of an experiment will provide necessary meta-data for a scientist to understand and recreate the results of an experiment. To support this we explored a model for the semantic description of a workflow-centric Research Object (RO), where an RO is defined as a resource that aggregates other resources, e.g., datasets, software, spreadsheets, text, etc. We have applied the workflow-centric RO model to bioinformatics and astronomy case studies. In Hettne et al 2014, three workflows were produced following defined Best Practices for workflow design.
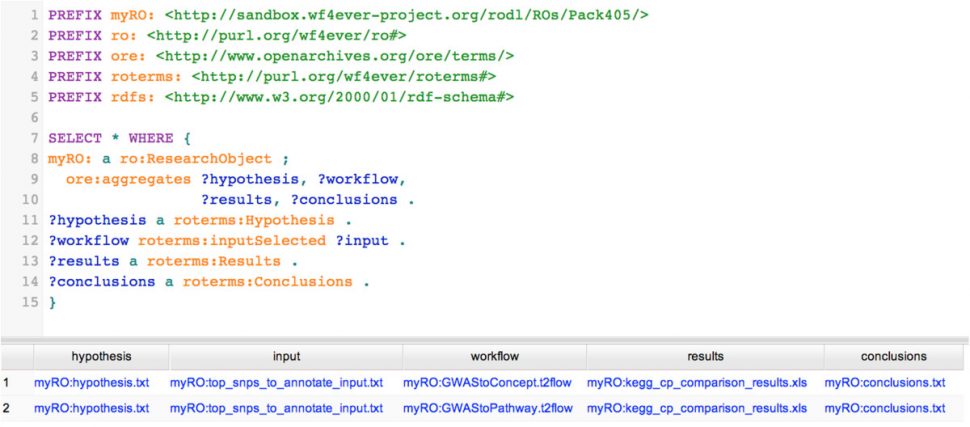
We applied this model to a case study where we analysed human metabolite variation by workflows. By modelling the experiment as an RO, we were able to automatically query the experiment and answer questions such as “which particular data was input to a particular workflow to test a particular hypothesis?”, and “which particular conclusions were drawn from a particular workflow?”.
Applying a workflow-centric RO model to aggregate and annotate the resources used in a bioinformatics experiment, allowed us to retrieve the conclusions of the experiment in the context of the driving hypothesis, the executed workflows and their input data. The RO model is an extendable reference model that can be used by other systems as well.